- Animal health
- Animal Nutrition
- Genetic and Biodiversity
- Environment and natural resources protection
- Socio-economy in pig production sector
- Quality and food safety
- Animal husbandry and sustainable practices
- Rural Development
2. The Institute of Animal Health, Pirbright Laboratory, Pirbright, Surrey, GU24 ONF, UK
African Swine Fever: virus isolates | Comparison of genomes of African swine fever virus isolates from Cameroon, other African countries and Europe
Determination of genetic relationship between ASF virus isolates from Cameroon, other African countries and Europe by restriction enzyme site mapping of their genomes.
Introduction
African swine fever (ASF) is an acute, highly contagious and often fatal disease of domestic pigs (17, 18). It is caused by a large cytoplasmically-located, icosaheral virus that contains a complex, linear double-stranded DNA genome (1, 7). Although morphologically similar to and originally classified with the Iridoviridae, more detailed analysis has revealed that ASF virus genome and replication resemble those of Poxviridae in many respects (1, 5), and the virus is at this time placed in a separate family (Asfaviridae) in which it is the only member. It has been established by restriction enzyme analysis and mapping restriction enzyme sites on genomes, that ASF virus isolates obtained from different parts of Cameroon at different times were very similar to each other (4, 10). It was shown in previous studies that genomes of virus isolates obtained from outbreaks in domestic pigs in Europe were closely related to each other (6, 24, 25), and that some European isolates were closely related to those from outbreaks in domestic pigs in the Caribbean and the isolate that caused the ASF epizootic in Cameroon in 1982 (25). The origin of some outbreaks in Europe were traced back to the Iberian peninsula on the basis of epizootiological information and evidence from restriction enzyme analysis and restriction enzyme site maps of the genomes of the virus isolates (3, 26). In contrast, other African isolates of ASF virus collected from various hosts in different geographical locations over a long time span were genetically diverse (4, 9, 22, 23, 25). However, all the African isolates used by these workers were obtained from the eastern and southern parts of Africa. Outbreaks of ASF have been reported in other parts of the continent such as Benin, Senegal, Guinea Bissau, Nigeria and Côte d’Ivoire in the West, Central African Republic, Chad, Congo, Democratic Republic of Congo, Sao Tome, Angola and Namibia in the Southwest (2, 23, 27).
This study was therefore carried out to determine the genetic relationship between ASF virus isolates from Cameroon, other African countries and Europe by restriction enzyme site mapping of their genomes.
Materials and methods

Table I: African swine fever virus isolates from Europe, Africa and Cameroon used in restriction enzyme analysis
The restriction enzymes, BamHI, Asp718 and XbaI obtained from Boehringer Mannheim, were used to digest the virus DNA following manufacturer’s recommendations. End-labeling of 32PdATP, using klenow fragment of DNA polymerase, was performed using standard procedures (20). Electrophoresis of the digested and end-labeled products was performed on 0.6% agarose (sigma type II) gel in 40 mM Tris-acetate buffer (pH 8.0). The gels were dried and the bands visualized by autoradiography. DNA was transferred from wet agarose gels onto Hybond-N filters (Amersham) by the method of Southern (21) and was covalently attached to the filters by heating in an oven at 80°C for 2 h. Hybridization was carried out using plasmid clones of the Vero cell-adapted Spanish isolate of ASF virus DNA as probes (19), and the procedure was as described by Feinberg and Vogelstein (13). Preparation of Southern filters, radioactively labeled probes and hybridization to Southern filters were all procedures used in the construction of restriction enzyme site maps of the ASF virus isolates of the study.
Results
Restriction enzyme analysis of the genomes of European and Cameroon ASF virus isolates
Three restriction enzymes, BamHI, Asp 718, and XbaI, were used to digest the DNA obtained from the various ASF virus isolates and the products analyzed by agarose gel electrophoresis. The results showed that the genomes of Cameroon ASF virus isolates were very similar to those of the European isolates used in the study (figures 1, 2 and 3), although one fragment (BamHI-L, Asp 718-I) varied in length between the isolates. CAM/86 ASF virus isolate also differed from other isolates in the mobility of the Asp 718-H and BamHI-0 fragment.
Comparison of BamHI restriction enzyme site maps of genomes of European and Cameroon isolates of ASFV

Figure 1: Restriction enzyme analysis of genomes of Cameroon and European isolates of the African swine fever virus using BamHI. Lane 1: Malta/78; lane 2: Sardinia/78; lane 3: Sardinia/ 82; lane 4: Italy/83; lane 5: Montijo (Lisbon)/84; lane 6: Belgium/ 85; lane 7: CAM/82; lane 8: CAM/85; lane 9: CAM/86; lane 10: CAM/87. Sizes of molecular weight markers in Kbp are indicated.
Cameroon and European ASFV isolates could be separated into four groups based on the variation in size of BamHI-L fragments even though their restriction enzyme fragment patterns were very similar (table III). Group 1 contained Belgium/85, CAM/82, CAM/85, CAM/87 and CAM/88 isolates. Group 2 included Montijo/84, Sardinia/78 and CAM/86, while Group 3 was made up of Malta/78, Sardinia/82 and Italy/83 isolates of ASFV.

The genome of the CAM/86 virus isolate contained an additional variation in the right terminal fragment which differed from the other members of Group 2, so it could actually be placed in a subgroup (Group 2a) within Group 2.
Restriction enzyme analysis of the genomes of ASFV isolates from Cameroon and other African countries
Virus DNA prepared from isolates from Cameroon, Senegal, Democratic Republic of Congo (DRC), Angola, Namibia, Malawi, Zambia and Mozambique were digested with the restriction enzyme BamHI, and the digested products were analyzed by agarose gel electrophoresis. The restriction enzyme fragment patterns of genomes of virus isolates from the east and southeast of Africa (Tanzania, Malawi, Zambia and Mozambique) were very different from those of the genomes of Cameroon virus isolates and were also different from each other (figures 5 and 6).
The genome of the Dakar/59 virus isolate from Senegal in West Africa was very similar to the genomes of southwest African virus isolates and also similar to the genomes of viruses from Cameroon (figures 5 and 6). The Dakar/59 and southwest African isolates were genetically very different from the east and southeast African virus isolates (figures 5 and 6).

Figure 5: Restriction enzyme analysis of genomes of African swine fever virus isolates from Europe, Cameroon and other African countries with the restriction enzyme BamHI. The figure shows positions and sizes of the molecular weight markers in Kbp. |
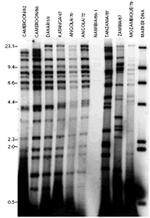
Figure 6: Restriction enzyme analysis of genomes of African swine fever virus isolates from Cameroon and other African countries with the enzyme BamHI. Molecular weight markers are indicated. |
Comparison of BamHI restriction enzyme site maps of genomes of Cameroon and African isolates of ASFV
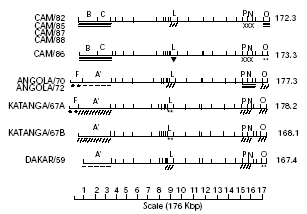
Figure 7: BamHI restriction enzyme site maps of virus genomes of Cameroon, Angola, Katanga/67 and Dakar/59 isolates of the African swine fever virus. The variable regions in each genome are indicated. |
No fragment size variations were observed within the BamHI-L fragment, which maps to the central region of the genome for DNA of CAM/82, CAM/85, CAM/87, CAM/88, Angola/70, Angola/72 and Dakar/59 virus isolates. Virus isolates from Cameroon with the exception of CAM/86 had BamHI-L fragments, which were the same in size as those of the genomes of the two viruses from Angola and the one from Senegal (figure 7). Variations were also observed in the Angola viruses, Katanga/67 and Dakar/59 isolates in the left terminal region, the region 10-35 Kb from the left terminus, the region about 10-20 Kb from the right terminus and the right terminus of the genome (figure 7). The right terminal fragment was the same in the genomes of Angola and Katanga/67 isolates, but differed from that of the Dakar/59 isolate (figure 7), and the difference was about 100 bp. The genomes of Katanga/67 and Dakar/59 virus isolates did not differ within the region about 10-20 Kb from the right terminus of the genome.
The virus genome of the Lil 20/1 isolate from Malawi (8) was very different from those from Cameroon and other isolates from the west and southwest of Africa by restriction enzyme site mapping, and it showed a marked difference in the order of the restriction enzyme fragments compared with the other virus isolates (8).
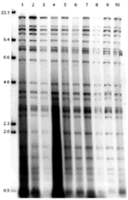
Figure 3: Restriction enzyme analysis of genomes of Cameroon and European isolates of the African swine fever virus with the restriction enzyme XbaI. Lanes 1-10 consist of ASFV isolates mentioned in the same order as in figure 1. Sizes of molecular weight markers in Kbp are indicated. |
Table II: Sizes of BamHI-L restriction enzyme fragments in genomes of isolates of African swine fever virus from Cameroon and Europe (in Kbp) 
Figure 4: Comparison of BamHI restriction enzyme site maps of genomes of Cameroon and European isolates of African swine fever virus. Variable regions in the different genomes are indicated. Restriction site maps of the genomes of virus isolates from Europe taken from Wilkinson et al. (unpublished results). Table III: Distribution of isolates of African swine fever virus from Cameroon and Europe into groups based on the sizes of BamHI-L restriction enzyme fragments 
|

Figure 8: Hybridization of the plasmid DNA clone RK’ with BamHI digests of CAM/82, CAM/86, Dakar/59, Katanga/67, Angola/72 and Lisbon/57 isolates of the African swine fever virus. Molecular weight markers in Kbp are indicated. |
Discussion
Restriction enzyme analysis of genomes of ASF virus isolates from Cameroon, other African countries and Europe showed that isolates from Cameroon and Europe were very closely related, except for the Lisbon/57 genome which had a longer left terminal region than the others. Also, the genomes of isolates from Angola, DRC, and Senegal were similar to those from Cameroon and Europe. Virus isolates from Angola and Katanga had a long left terminal left region similar in size to the Lisbon/57 isolates. Isolates from East and Southeast Africa were genetically different from Cameroon and European isolates and also different from each other. Comparison of BamHI restriction enzyme site maps of the virus genomes of the Cameroon and European isolates showed a variation in length of the BamHI-L fragment, which maps to the middle of the genome (figure 4). By comparing the variation in the BamHI-L fragment size, Cameroon and European isolates were separated into four groups: Group 1 contained the Belgium/85, CAM/82, CAM/85, CAM/87 and CAM/88 isolates, while Group 2 had the Montijo/84 and Sardinia/78 isolates with a subgroup 2a containing the CAM/86 isolate only. The CAM/86 virus isolate was put into a subgroup (Group 2a) because of the additional difference in size of the right terminal fragment despite the equality in size of the BamHI-L fragment with the other members of Group 2. Group 3 was made up of the Malta/78, Sardinia/82 and Italy/83 virus isolates (tables II and III).
Other workers have reported similarity in the restriction enzyme fragment patterns of genomes of ASF virus isolates obtained from outbreaks in domestic pigs in Europe, the Caribbean and the 1982 ASF outbreak in Cameroon (24, 25), and the results presented in this study were similar to their findings about the genomes of isolates from Cameroon and Europe. Although the Lisbon/57 virus isolate showed a similar restriction enzyme fragment pattern to those of the genomes of the Cameroon isolates, differences in fragment sizes were observed at both ends of the genome (figure 4); it had an additional 6 Kb sequence at a position about 10 Kb from the left terminus (24).
A comparison of the genomes of ASF isolates from Cameroon with those of isolates from other African countries by restriction enzyme analysis showed that the virus isolates from Senegal (Dakar/59), DRC (Katanga/67), Angola (Angola/70, Angola/72) and Namibia (Namibia/86-1) were all similar to the genomes of Cameroon and European ASFV isolates (figure 5). The genomes of virus isolates from East and Southeast Africa were genetically very different from isolates from Cameroon, Europe, West and Southwest African countries, and were also very different from each other (figures 5 and 6). The RK’ plasmid clone which contains DNA from the left terminus of the BA71V Spanish isolate (19) hybridized to two fragments in the genome of the Katanga/67 virus isolate, BamHI-A’ and BamHI-F (figure 8). This resulted in the construction of two alternative restriction enzyme site maps for the genome of the Katanga/67 ASFV isolate (figure 7). Two possible explanations could be given for this phenomenon.
First, it is possible that the virus DNA was extracted from two virus populations in the Katanga/67 virus isolate which differed in the left terminal fragment of their genomes as observed by the crosshybridization with the RK’ plasmid clone (figure 8). In the Katanga/67B virus population, a deletion may have occurred near the end of the genome which was the same size as the BamHI-F fragment and also removed the Sal I site between fragments F and A’. The genome map would therefore have a left terminal fragment the same length at the A’ fragment and no F fragment (figure 7). The Katanga/67A virus population would have the BamHI-F fragment as the left terminal fragment since the F fragment in the Katanga/67B DNA was of reduced intensity compared to those in other virus isolates (figure 8). This indicates that the F fragment is present in a reduced molar ratio supporting the hypothesis that two virus populations were present in the Katanga isolate. The other possible explanation is that the Katanga/67 virus probably has sequences in the BamHI-A’ fragment which were either the 360 multigene family or tandem repeats closely related to those in the terminal inverted repetitions as observed in the BA71V strain of ASFV (14), and these hybridize with the RK’ plasmid DNA clone. The Lisbon/57 and Angola viruses did not show such hybridization with the RK’ plasmid clone, hence they do not have these sequences within the BamHI-A’ fragment. However, a comparison of restriction enzyme site maps of genomes of Cameroon and west and southwest African isolates showed that there were variations in the central region and in both termini of the genomes (figure 7). The order of restriction enzyme fragments was the same only for the virus isolates from Angola and the Katanga/67A, and the same for the Katanga/67B and Dakar/59 isolates (figure 7). The order of restriction enzyme fragments of the genome of the Lil20/1 isolate from Malawi (8) was very different from those of the other virus isolates. However, Dixon (8) observed that some BamHI fragments were conserved or were similar in length both in the Lil 20/1 and European isolates. These fragments were generally located in the right terminal 50 Kb, indicating that this region contained (by comparison) the most conserved sequences of the genome.
The genetic diversity of ASF virus isolates from the east and southeast of Africa has been reported by other workers (3, 4, 9, 22). It is also known that in these areas the virus is present in warthogs, soft ticks and domestic pigs (3, 4, 15, 16, 27). It is therefore possible that the genetic diversity observed in virus isolates from these areas may reflect the selective pressure imposed by the genetically different hosts in the area. On the other hand, the similarity of isolates from domestic pigs in Cameroon, Europe, the west and southwest of Africa indicated that all these domestic pig isolates may have originated from a single introduction of the ASF virus from a wildlife source into the domestic pig population. Once introduced into this population the virus was readily transmitted between pigs. The results of this study have shown that the persistent outbreaks of ASF in domestic pig populations in Cameron may have been due to the reintroduction of closely related virus isolates from infected areas outside the country. The possible role of soft ticks and warthogs in the transmission and persistence of the disease in the three main pig-producing provinces of Cameroon had also been investigated (11) and there was no evidence of the presence of these reservoir hosts of ASFV in these provinces.
References
- ALMENDRAL J.M., BLASCO R., LEY V., BELOSO A., TALAVERA A., VINUELA E., 1984. Restriction site map of African swine fever virus DNA. Virology, 133: 258-270.
- ANIMAL HEALTH YEAR BOOK, 1987. Rome, Italy, FAO, Animal Production & Health Division, p. 160.
- ANNUAL REPORT OF THE INSTITUTE FOR ANIMAL HEALTH, 1994. Berks, UK, Compton, p. 132.
- ANNUAL REPORT OF THE INSTITUTE FOR ANIMAL HEALTH, 1996. Berks, UK, Compton, p. 140.
- BAROUDY B.M., VENKATESAN S., MOSS B., 1982. Incomplete basepaired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterupted polymicleotide chain. Cell, 28: 315-324.
- BLASCO R., AGUERO M., ALMENDRAL J.M., VINUELA E., 1989. Variable and constant regions in African swine fever virus DNA. Virology, 168: 330-338.
- CARRASCOSA J.L., CARAZO J.M., CARRASCOSA A.L., GARCIA N., SANTISTEBAN A., VINUELA E., 1984. General morphology and capsid fine structure of African swine fever virus particles. Virology, 132: 160-172.
- DIXON L.K., 1988. Molecular cloning and restriciton enzyme mapping of an African swine fever virus isolate from Malawi. J. gen. Virol., 69: 1683-1694.
- DIXON L.K., WILKINSON P.J., 1988. Genetic diversity of African swine fever virus isolates from soft ticks (Ornithodoros moubata) inhabiting burrows in Zambia. J. gen. Virol., 69: 2981-2993.
- EKUE N.F., 1989. PhD Thesis, University of Surrey, Guildford, UK, 231 p.
- EKUE N.F., WILKINSON P.J., 1990. Absence of Ornithodoros moubata, the vector of African swine fever virus, from the main pig producing area of Cameroon. Trop. Anim. Health Prod., 22: 127-131.
- EKUE N.F., WILKINSON P.J., 1999. Analysis of the genomes of African swine fever virus isolates from Cameroon. Revue Elev. Méd. vét. Pays trop., 52: 195-201.
- FEINBERG A.P., VOGELSTEIN B., 1983. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem., 132: 6-13.
- GONZALEZ A., CALVO V., ALMAZAN F., ALMENDRAL J.M., RAMIREZ J.C., DELA VEGA I., BLASCO R., VINUELA E., 1989. Multigene family in African swine fever virus DNA. Family 360. J. mol. Biol. In: J. Virol., 64: 2073-2081.
- HARESNAPE J.M., MANU F.D., 1986. The distribution of ticks of the Ornithodoros moubata complex (Ixodoidea: Argasidae) in Malawi and its relation to African swine fever epizootiology. J. Hyg., 96: 535-544.
- HARESNAPE J.M., WILKINSON P.J., MELLOR P.S., 1988. Isolation of African swine fever virus from ticks of the Ornithodoros moubata complex (Ixodoidea Argasidae) collected within the African swine fever enzootic area of Malawi. Epidemiol. Infect., 101: 173 185.
- HESS W.R., 1971. ASFV. Virology monographs, 9: 1-33. New York, NY, USA, Springer-Verlag.
- HESS W.R., 1981. African swine fever: a reassessement. Adv. vet. Sci., 25: 39-69.
- LEY V., ALMENDRAL J.M., CARBONERO P., BELOSO A., VINUELA E., TALAVERA A., 1984. Molecular cloning of African swine fever virus DNA. Virology, 133: 249-257.
- MANIATIS T., FRITSCH E.F., SAMBROCK J., 1982. Molecular cloning: A laboratory manual. New York, NY, USA, Cold Spring Habor Laboratory, 342 p.
- SOUTHERN E.M., 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. mol. Biol., 9: 503-518.
- SUMPTION K.J., HUTCHINGS G.H., WILKINSON P.J., DIXON L.K., 1990. Variable regions on the genome of the Malawi isolate of African swine fever virus. J. gen. Virol., 71: 2331-2340.
- THOMSON G.R., 1985. The epidemiology of African swine fever: the role of free-living hosts in Africa. Oonderst. J. vet. Res., 52: 201-209. 24. VINUELA E., 1985. African swine fever. Curr. Top. Microbiol. Immunol., 116: 151-170.
- WESLEY R.D., TUTHILL A.E., 1984. Genome relatedness among African swine fever virus field isolates by restriction endonuclease analysis. Prev. vet. Med., 2: 53-62.
- WILKINSON P.J., 1986. Epidemiology of African swine fever. Revue sci. tech. Off. int. Epizoot., 5: 487-493.
- WILKINSON P.J., PEGRAM R.G., PERRY B.D., LEMCHE J., SCHELS H.F., 1988. The distribution of African swine fever virus isolated from Ornithodoros moubata in Zambia. Epidemiol. Infect., 101: 547-564
For further information


Comments
CIRAD © 2007 (All rights reserved) - Disclaimer stating - Page updated : 12/04/2007
